EXPERIMENT OBJECTIVE
In this investigation, students will:
1. Create cladograms that depict evolutionary relationships among organisms.
2. Analyze biological data with a sophisticated bioinformatics online tool.
3. Connect and apply concepts pertaining to genetics and evolution.
In this investigation, students will:
1. Create cladograms that depict evolutionary relationships among organisms.
2. Analyze biological data with a sophisticated bioinformatics online tool.
3. Connect and apply concepts pertaining to genetics and evolution.
Background information:
The genetic revolution will forever contribute new discoveries. While scientist continue to identify genes that cause disease or phenotypic differences, there is a growing danger to see humans merely as a sum of their genes; comprehending the ethical, legal, and social outcomes of genetic knowledge, and the development of policy options for public consideration are therefore yet another major part of the human genome research effort. This simplistic view may create situations in which genetic information has the potential to cause inconvenience or harm. Data from DNA sequencing is limited unless it can be changed to biologically useful information. Bioinformatics therefore is a critical part of DNA sequencing . It changed from the merging of computer technology and biotechnology. The widespread use of the internet has made it possible to easily retrieve information from the various genome projects. These exercises will involve using BLASTN will compare a nucleotide sequence and other sequences in the nucleotide database and will also compare amino acid sequences with other amino acid sequences.
Investigation I:
EXERCISE 1:
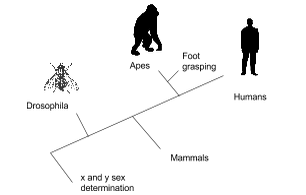
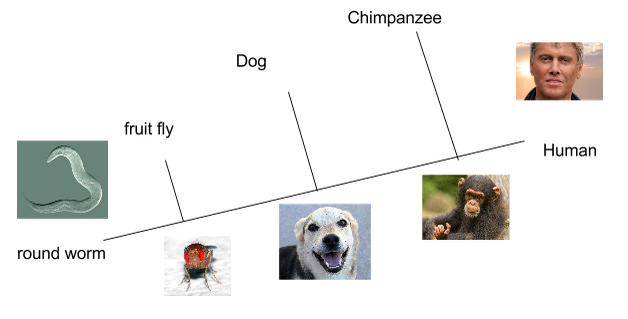
Chimpanzees and humans share 96% of their DNA which would place them closely on a cladogram. Humans and fruit flies are placed farther apart on a cladogram since they share only approximately 60% of their DNA. In the space provided, draw a cladogram that shows the evolutionary relationship between humans, chimpanzees, and fruit flies.
EXERCISE 2:
The genetic revolution will forever contribute new discoveries. While scientist continue to identify genes that cause disease or phenotypic differences, there is a growing danger to see humans merely as a sum of their genes; comprehending the ethical, legal, and social outcomes of genetic knowledge, and the development of policy options for public consideration are therefore yet another major part of the human genome research effort. This simplistic view may create situations in which genetic information has the potential to cause inconvenience or harm. Data from DNA sequencing is limited unless it can be changed to biologically useful information. Bioinformatics therefore is a critical part of DNA sequencing . It changed from the merging of computer technology and biotechnology. The widespread use of the internet has made it possible to easily retrieve information from the various genome projects. These exercises will involve using BLASTN will compare a nucleotide sequence and other sequences in the nucleotide database and will also compare amino acid sequences with other amino acid sequences.
Investigation I:
EXERCISE 1:
Chimpanzees and humans share 96% of their DNA which would place them closely on a cladogram. Humans and fruit flies are placed farther apart on a cladogram since they share only approximately 60% of their DNA. In the space provided, draw a cladogram that shows the evolutionary relationship between humans, chimpanzees, and fruit flies.
EXERCISE 2:
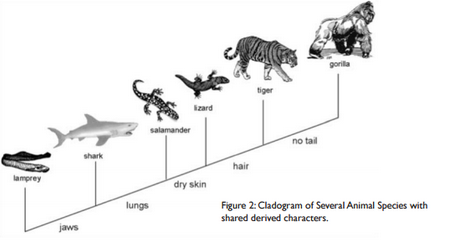
Question 1: According to the cladogram, what organisms have hair?
Tigers and Gorillas have hair according to cladogram.
Question 2: According to the cladogram, what four structures do tigers possess?
Tigers have jaws, lungs, dry skin, and hair.
Question 3: According to the cladogram, which structure evolved first between lungs and dry skin?
Lungs evolved first compared to dry skin.
Tigers and Gorillas have hair according to cladogram.
Question 2: According to the cladogram, what four structures do tigers possess?
Tigers have jaws, lungs, dry skin, and hair.
Question 3: According to the cladogram, which structure evolved first between lungs and dry skin?
Lungs evolved first compared to dry skin.
Investigation II:
Exercise 3:
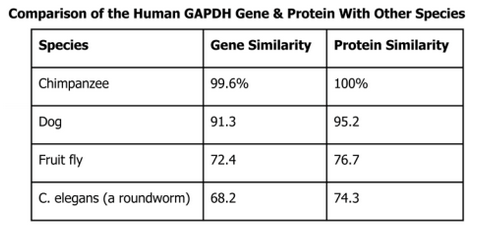
Use the data below to create a cladogram
Use the data below to create a cladogram
Step 1: Instruction for Downloading Gene Files
Before starting this investigation, students need to download four gene files (Gene
1- Gene 4) to their fl ash drive or computer. These fi les are located at the following web
address: http://blogging4biology.edublogs.org/2010/08/28/college-board-lab-files/
Note that these files will not open on your computer. They only work when opened on
the BLAST website.
Step 2: Instructions for BLAST Queries
Upload the gene sequences into BLAST by following the instructions below:
1. Go to the BLAST homepage: http://blast.ncbi.nlm.nih.gov/Blast.cgi
2. Click on “Saved Strategies” on the top of the page.
3. Under “Upload Search Strategy,” click on “Browse” and locate one of the gene files you saved onto your computer.
4. Click “View.”
5. A screen will appear with the parameters for your query already configured.
NOTE: Do not alter any of the parameters. Scroll down the page and click on the “BLAST” button at the bottom.
6. After collecting and analyzing all of the data for that particular gene (see instructions below), repeat this procedure for the other three gene sequences.
Step 3: Instructions for Analyzing BLAST Queries
1. Scroll down to the section titled, “Sequences producing significant alignments”. The list of organisms that appear below this section are those with sequences identical to or most similar to the gene of interest. The most similar sequences are listed first and as you move down the list, the sequences become less similar to your gene of interest.
2. Try clicking on a particular species listed to get a full report that includes the species’ classification scheme, the research journal the gene was first reported in, and the sequence of bases that appear to align with your gene of interest.
3. Click “Distance tree of results,” a cladogram of the species with similar sequences to your gene of interest placed on the cladogram will be shown according to how closely their matched gene aligns with your gene of interest.
Step 4: Analysis of Results
Species share similar genes because of common ancestry. The more similar genes two spe-
cies have in common, the more recent their common ancestor. Thus, the two species will be located closer on a cladogram.
As you collect information from BLAST for each of the gene files explain whether the data supports your original hypothesis and your original placement of the fossil species on the cladogram.
Step 5: Drawing the cladogram
Redraw the original cladogram and include your final placement of the fossil species.
Before starting this investigation, students need to download four gene files (Gene
1- Gene 4) to their fl ash drive or computer. These fi les are located at the following web
address: http://blogging4biology.edublogs.org/2010/08/28/college-board-lab-files/
Note that these files will not open on your computer. They only work when opened on
the BLAST website.
Step 2: Instructions for BLAST Queries
Upload the gene sequences into BLAST by following the instructions below:
1. Go to the BLAST homepage: http://blast.ncbi.nlm.nih.gov/Blast.cgi
2. Click on “Saved Strategies” on the top of the page.
3. Under “Upload Search Strategy,” click on “Browse” and locate one of the gene files you saved onto your computer.
4. Click “View.”
5. A screen will appear with the parameters for your query already configured.
NOTE: Do not alter any of the parameters. Scroll down the page and click on the “BLAST” button at the bottom.
6. After collecting and analyzing all of the data for that particular gene (see instructions below), repeat this procedure for the other three gene sequences.
Step 3: Instructions for Analyzing BLAST Queries
1. Scroll down to the section titled, “Sequences producing significant alignments”. The list of organisms that appear below this section are those with sequences identical to or most similar to the gene of interest. The most similar sequences are listed first and as you move down the list, the sequences become less similar to your gene of interest.
2. Try clicking on a particular species listed to get a full report that includes the species’ classification scheme, the research journal the gene was first reported in, and the sequence of bases that appear to align with your gene of interest.
3. Click “Distance tree of results,” a cladogram of the species with similar sequences to your gene of interest placed on the cladogram will be shown according to how closely their matched gene aligns with your gene of interest.
Step 4: Analysis of Results
Species share similar genes because of common ancestry. The more similar genes two spe-
cies have in common, the more recent their common ancestor. Thus, the two species will be located closer on a cladogram.
As you collect information from BLAST for each of the gene files explain whether the data supports your original hypothesis and your original placement of the fossil species on the cladogram.
Step 5: Drawing the cladogram
Redraw the original cladogram and include your final placement of the fossil species.
Investigation III:
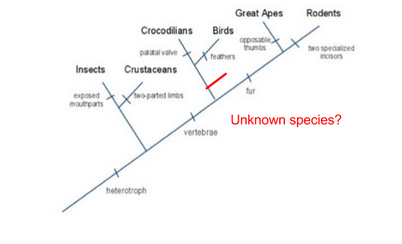
Hypothesis: If the organisms shares most of its DNA with the common bird then it could have once been a dinosaur. Observations: Organism has a vertebrate, has a tail constructed of bone, and has 2 legs that are clearly visible. Smaller body frame. Pointed towards the mouth area. Tail is very long, probably same length as body, if not even more.
Initial Cladogram:
Blast Results:
Questions:
1. What species has the most similar gene sequence as your gene of interest?
2. Where is that species located on the cladogram?
3. How similar is that gene sequence?
4. What species has the least similar gene sequence as your gene interest?
When studying all 4 of the genetic trees, 2 out 4 show that the fossil was connected with the birds family overall compared to every other family. They shared the most genes with them. The species should be located on the alligator/bird branch. This is because, although it is related to the bird, it is also partially related to the Crocodilian family. The gene sequence must be very close because the fossil goes through lineages and lineages of bird lines, making it very close to the avian family. On the flip side, the species with the least similar gene sequence as the fossil would be rodents. The fossil has some ancestory shared with the flies so it makes the right side of the cladogram less similar.
2. Where is that species located on the cladogram?
3. How similar is that gene sequence?
4. What species has the least similar gene sequence as your gene interest?
When studying all 4 of the genetic trees, 2 out 4 show that the fossil was connected with the birds family overall compared to every other family. They shared the most genes with them. The species should be located on the alligator/bird branch. This is because, although it is related to the bird, it is also partially related to the Crocodilian family. The gene sequence must be very close because the fossil goes through lineages and lineages of bird lines, making it very close to the avian family. On the flip side, the species with the least similar gene sequence as the fossil would be rodents. The fossil has some ancestory shared with the flies so it makes the right side of the cladogram less similar.
Investigation IV:
The gene I chose was ACTC1. "The protein encoded by this gene belongs to the actin family which is comprised of three main groups of actin isoforms, alpha, beta, and gamma. The alpha actins are found in muscle tissues and are a major constituent of the contractile apparatus."(NCBI) This protein found in eukaryotic cells make up the main part of a cell's cytoskeleton. Actin and myosin makeup the skeletal fibers. They are what's responsible for muscle contractions and relaxations. They pretty much help us use our muscles. Other organisms are of course going to carry this gene or else they wouldn't be able to move around or contract their muscles.


 RSS Feed
RSS Feed
